TCR
BCR
The term “specific immunity receptors” includes the TCR (T-Cell Receptor) and the BCR (B-Cell Receptor), respectively the specific antigen receptors carried by T cells and B cells. Like a large majority of receptors, they are transmembrane molecules whose goal is to induce a modification in the cellular program through downstream intracellular molecular signalling pathways.
TCR and the BCR have many common features, but also major differences.
Common features
Antigen specificity
Antigen specificity is in essence the characteristic of adaptive immunity, which differentiates it from innate immunity. In other words, the receptor-ligand interaction must be absolutely specific in order to cause the engagement of the TCR or BCR.
Molecular structure
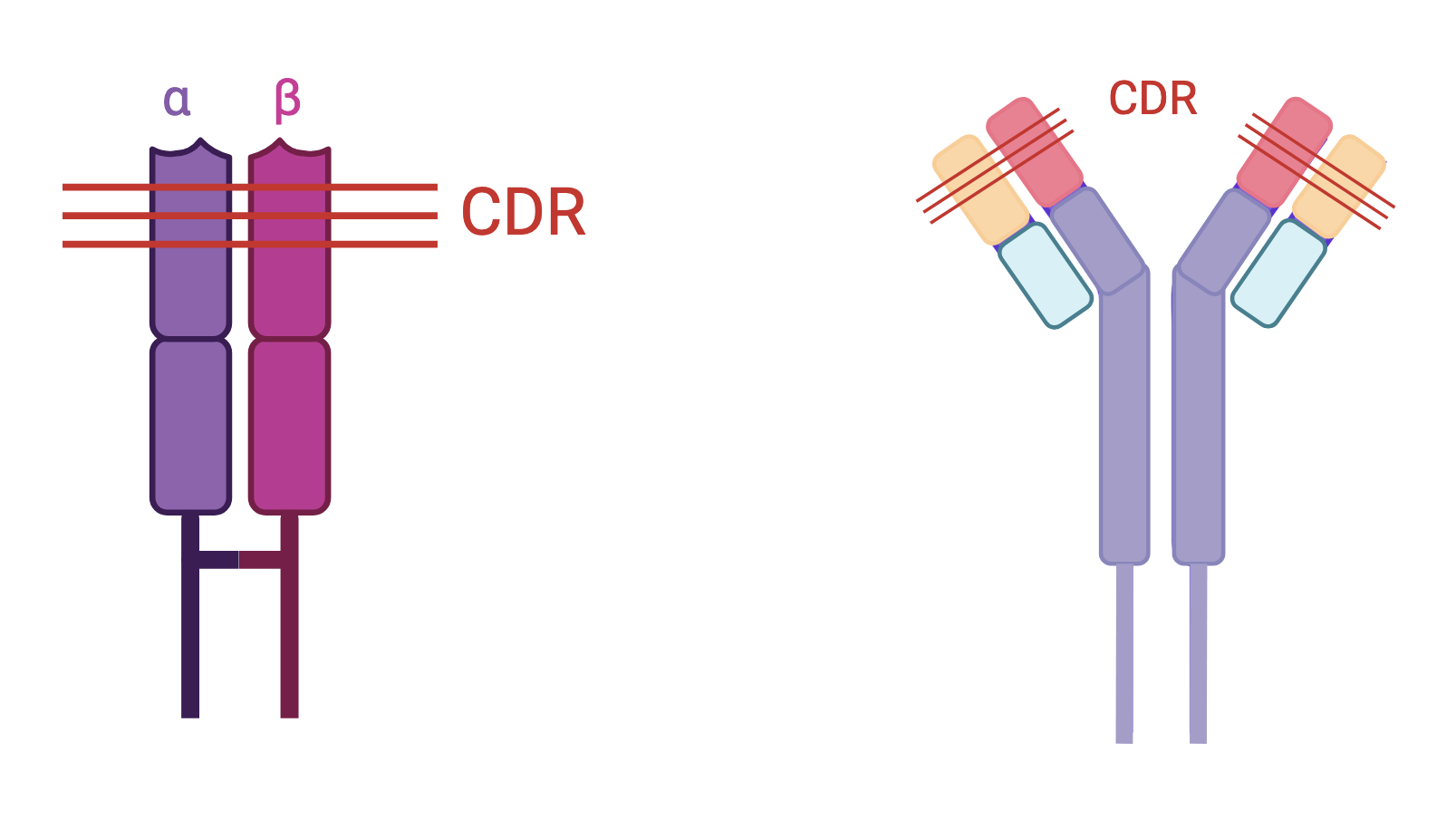
Although the product of distinct genes, TCR and BCR have a similar tertiary structure, both belonging to the immunoglobulin superfamily. They each have:
- An antigen recognition module, itself divided into constant (C) and variable (V) regions. The variable regions are located at the end of the receptors, they support antigen recognition, and they are called paratopes.
The invariability of constant regions makes it possible to preserve a common entry into downstream molecular pathways.
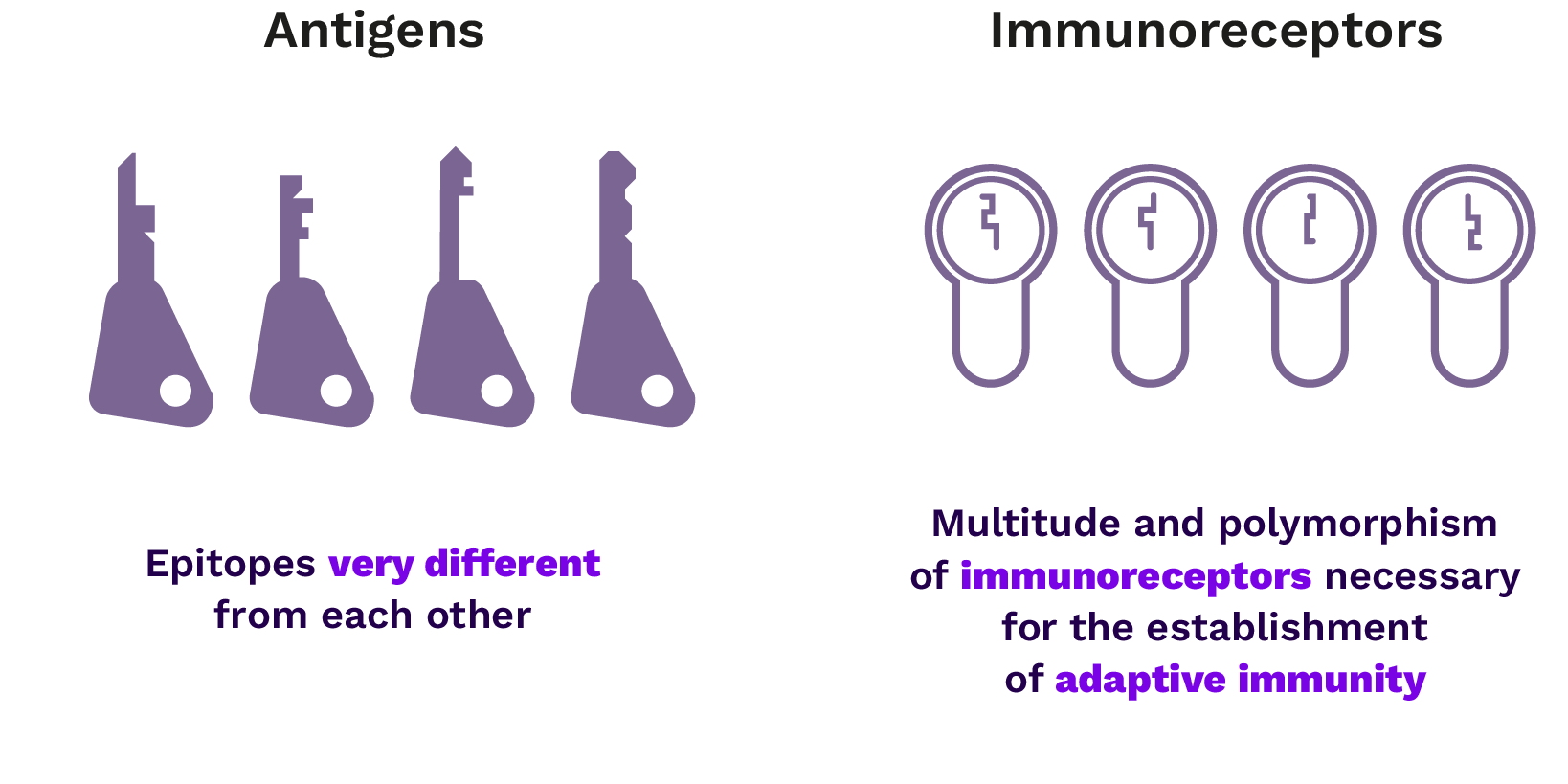
Polymorphism of the antigen recognition module
Given the infinite diversity of antigens and taking into account the specificity of the interaction between these antigens and the receptors of adaptive immunity, the polymorphism of the TCR and BCR must also be infinite.
However, how could lymphocytes produce an infinite number of different receptors, when – like all other nucleated cells in the body – their genome only contains a finite number of genes? This diversity is made possible by somatic rearrangements of genes encoding these receptors, a mechanism known as “V(D)J recombination”.
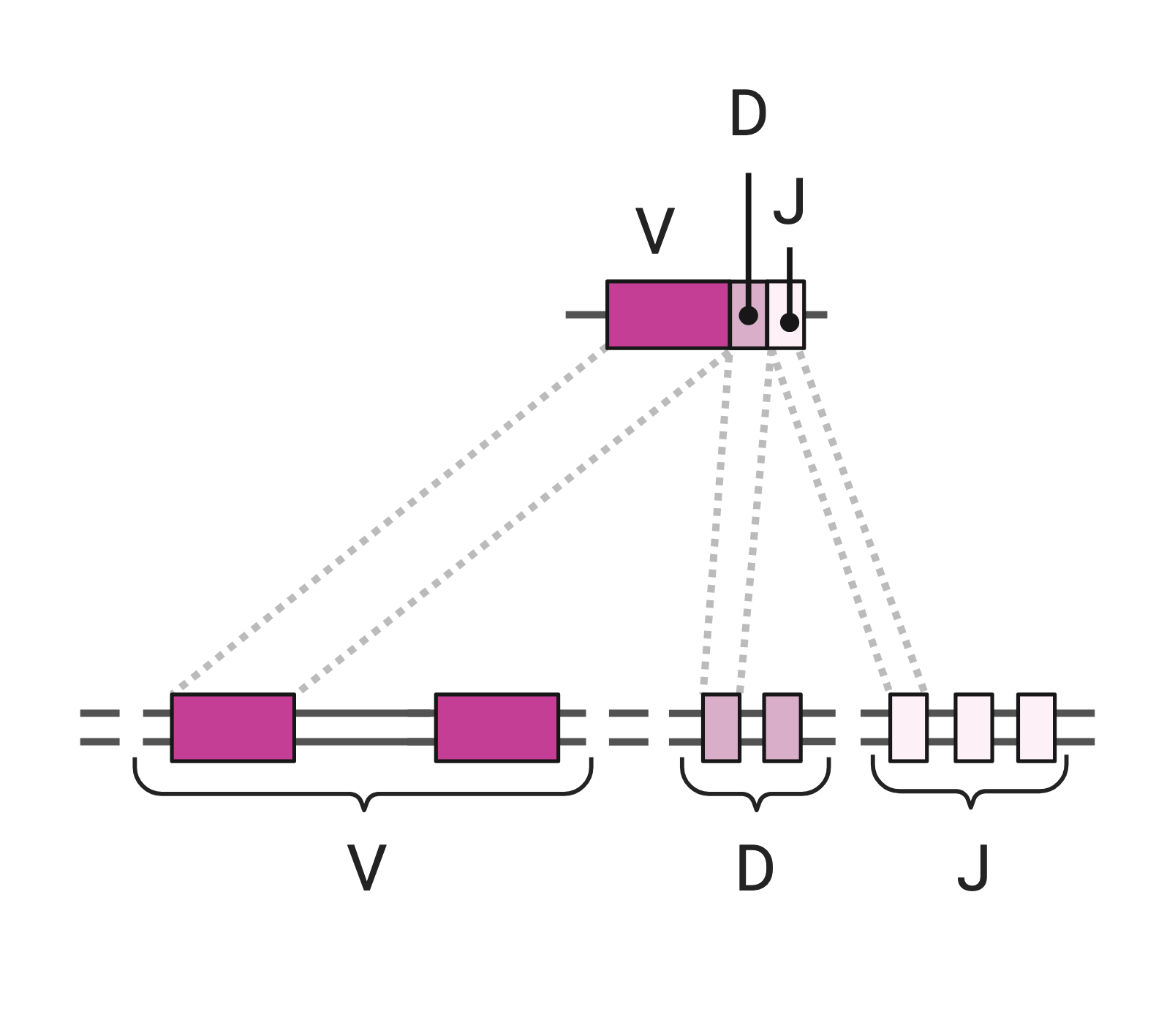
The mechanism is similar for the TCR and the BCR, in particular involving the same actors to generate two types of diversity:
- combinatorial diversityresulting from the random combination of a D gene with a J gene to form a DJ fragment, and secondly of the V gene with the DJ fragment to form a VDJ fragment. When the D fragment is absent (κ/λ light chains of the BCR and the α TCR), recombination takes place in one step with the random association of a V gene with a J gene to form a VJ fragment.
- junctional diversity, i.e. the addition and modification of nucleotides at rearrangement points.
V(D)J recombination is finely regulated by enzymes specific to the lymphocyte lineage, in particular:
- RAG (Recombination Activating Genes) 1 and 2, which ensure combinatorial diversity.
- TdT (Terminal Deoxynucleotidyl-Transferase), which ensures junctional diversity.
The recombination of the V(D)J gene segments will thus encode the variable regions of the TCR and BCR. These variable regions themselves include hypervariable regions corresponding to the ultra-specific region of the paratope (CDR Complementarity Determining Region) and so-called framework regions (FR Framework Region) which are paradoxically very conserved.
For more details on V(D)J recombination of the TCR and BCR, see the dedicated chapters: TCR and BCR.
Selection of immunoreceptors in lymphoid organs
Given that somatic recombination responsible for immunoreceptor polymorphism is a completely random mechanism, it is necessary to select the lymphocytes generated in order toprevent them from recognising autoantigens and causing autoimmune reactions. Selection takes place in the lymphoid organs (bone marrow, lymph nodes , and spleen for the BCR ; thymus for the TCR) (see T cell ontogeny / B cell ontogeny).
Antigen recognition/signal transduction decoupling
The diversity induced by V(D)J recombination is concentrated in the variable region of the receptor which corresponds to the paratope, that is to say, the part of the immunoreceptor specifically recognising the antigen. Thus, the constant regions are preserved, which makes it possible to preserve a common entry into the downstream molecular pathways.
Downstream pathways
The recognition of the specific antigen of the immunoreceptor of adaptive immunity induces the rapprochement of receptors and co-receptors and a reorganisation of the underlying cell membrane, leading to the creation of lipid rafts, the starting point of a molecular cascade, which will activate three pathways:
- Ras - MAPkinases - AP1
- Calmodulin - calcineurin - NFAT
- NFKB
Key differences
| TCR | BCR | ||
|---|---|---|---|
| Receptor | Cell | T cell | B cell |
| Description | Heterodimer | Heterotetramer: 2 heavy chains and 2 identical light chains 2 by 2 | |
| Architecture | αβ or γδ | 2 heavy chains (μ or δ) 2 light chains (κ or λ) | |
| Selection see T cell ontogeny and B cell ontogeny) | Thymus: Positive selection and negative selection | Bone marrow (central tolerance) Negative selection, BCR editing or cell anergy Blood, spleen, and lymph nodes (peripheral tolerance) Negative selection, BCR editing or cell anergy | |
| Maturation after V(D)J recombination | No | Somatic hypermutation of variable regions after encounter with antigen Class switching: change of the constant region and removal of the transmembrane and intracytoplasmic part during B cell > plasma cell transformation | |
| Valence | Univalent (single paratope) | Multivalent (2 paratopes) | |
| Recognised antigen | Antigen type | Peptides of some amino acids | Whatever |
| Primer | In HLA molecules: Intracellular antigen (Class I HLA) Extracellular antigen (Class II HLA (and I for dendritic cell (cross-presentation)) | Native = no primer (recognised directly by BCR) Obligatory extracellular | |
| Co-receptors | Signal transduction | CD3 | CD79 |
| Stabilisation of the interaction | CD4 or CD8 | CD21 (optional commitment) |